Phylogenomics resolve evolutionary relationships and providefirst insights into floral evolution in the tribe Shoreeae (Dipterocarpaceae)
- Background and Aims: A supra-annual, community-level synchronous flowering prevails in several parts of the tropical forests of Southeast Asia and its evolution has been hypothesized to be linked to pollinator shifts. The aseasonal Southeast Asian lowland rainforests are dominated by Dipterocarpaceae, which exhibit great floral diversity, a range of pollination syndromes and include species with annual and supra-annual gregarious flowering. Phylogenetic relationships within this family are still unclear, especially in the tribe Shoreeae.
- Methods: Here, we develop a pipeline to maximize recovery of genome-wide SNPs from restriction-site associated DNA sequencing (RADseq) in non-model organisms across wide phylogenetic scales. We then infer phylogenomic relationships in the tribe Shoreeae using both traditional and coalescent analyses.
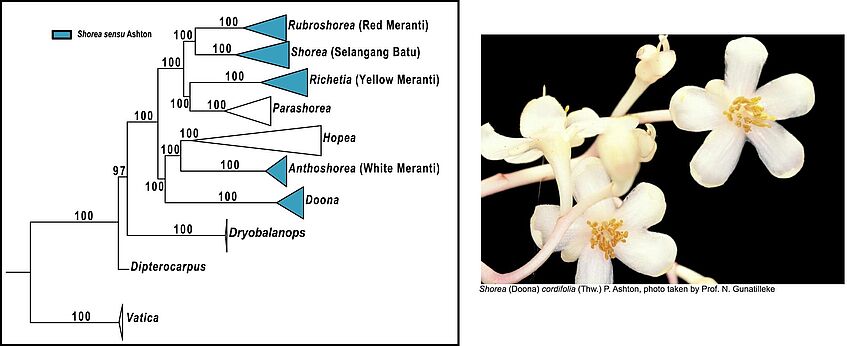
- Key Results: The phylogenetic trees obtained with these methods are congruent to each other and highly resolved. They allow reconstructing the evolutionary patterns of floral traits (number of stamens, anther structure and anther/appendage size) in the group. Our inferences indicate that species with many stamens, but smaller, globose anthers and longer appendages and have evolved multiple times from species with fewer stamens, but larger, oblong anthers and shorter appendages.
- Conclusions: This could have happened in parallel to iterative shifts in pollinators across the uncovered phylogeny from larger, longer generation to smaller, shorter-generation insects that can quickly build up the necessary population sizes during mass flowering episodes.